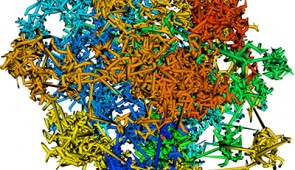
Chromatin organizes itself into 3D ‘forests’ in single cells
Knowledge could help scientists understand chromatin’s role in cancer, other diseases
- Link to: Northwestern Now Story
A single cell contains the genetic instructions for an entire organism. This genomic information is managed and processed by the complex machinery of chromatin -- a mix of DNA and protein within chromosomes whose function and role in disease are of increasing interest to scientists.
A Northwestern University research team -- using mathematical modeling and optical imaging they developed themselves -- has discovered how chromatin folds at the single-cell level. The researchers found chromatin is folded into a variety of tree-like domains spaced along a chromatin backbone. These small and large areas are like a mixed forest of trees growing from the forest floor. The overall structure is a 3D forest at microscale.
Chromatin is responsible for packing DNA into the cell nucleus. In humans, that’s about six feet of DNA in each cell. The new work suggests that chromatin is more structured and hierarchical in single cells than previously thought. Learning how chromatin correctly operates will help scientists understand what goes wrong with it in cancer and other diseases.
“By integrating theoretical and experimental work, we have produced a new chromatin folding picture that helps us see how the 3D genome is organized at the single-cell level,” said Igal Szleifer, the Christina Enroth-Cugell Professor of Biomedical Engineering at Northwestern’s McCormick School of Engineering. He co-led the research team with Vadim Backman.
Details of the interdisciplinary study were published today (Jan. 10) in the journal Science Advances.
“If genes are the hardware, chromatin is the software,” said Backman, the Walter Dill Scott Professor of Biomedical Engineering and director of the Center for Physical Genomics and Engineering. “If the structure of chromatin changes, it can alter the processing of the information stored in the genome, but it does not alter the genes themselves. Understanding chromatin folding holds the key to understanding how cells differentiate and how cancer happens.”
Advances in genomic, imaging and information technologies are just beginning to enable scientists to better understand how chromatin works. The Northwestern researchers used a Partial Wave Spectroscopic (PWS) microscope, optical imaging developed by Backman and colleagues, to peer deep into live cells and “sense” alterations in chromatin packing. They also used electron imaging.
“Our paradigm-shifting picture of chromatin folding is an important missing piece in the holistic view of genomic structure,” said Kai Huang, the study’s first author. Huang is a postdoctoral fellow in Szleifer’s research group. “The results should inspire new strategies to fight cancer.”
The research was supported by the National Science Foundation and the National Cancer Institute at the National Institutes of Health.
The paper is titled “Physical and data structure of 3D genome.” The corresponding authors are Szleifer, Backman and Huang.
Backman also is professor of medicine and of biochemistry and molecular genetics at the Feinberg School of Medicine, associate director for research technology and infrastructure at the Robert H. Lurie Comprehensive Cancer Center of Northwestern University and a member of the Chemistry of Life Processes Institute.
Szleifer is also a member of the Lurie Cancer Center.
Multimedia Downloads
Chromatin forests
Please credit all images to Northwestern University